Evaluating coverage bias in next-generation sequencing of Escherichia coli
4.7 (585) In stock
4.7 (585) In stock
Whole-genome sequencing is essential to many facets of infectious disease research. However, technical limitations such as bias in coverage and tagmentation, and difficulties characterising genomic regions with extreme GC content have created significant obstacles in its use. Illumina has claimed that the recently released DNA Prep library preparation kit, formerly known as Nextera Flex, overcomes some of these limitations. This study aimed to assess bias in coverage, tagmentation, GC content, average fragment size distribution, and de novo assembly quality using both the Nextera XT and DNA Prep kits from Illumina. When performing whole-genome sequencing on Escherichia coli and where coverage bias is the main concern, the DNA Prep kit may provide higher quality results; though de novo assembly quality, tagmentation bias and GC content related bias are unlikely to improve. Based on these results, laboratories with existing workflows based on Nextera XT would see minor benefits in transitioning to the DNA Prep kit if they were primarily studying organisms with neutral GC content.
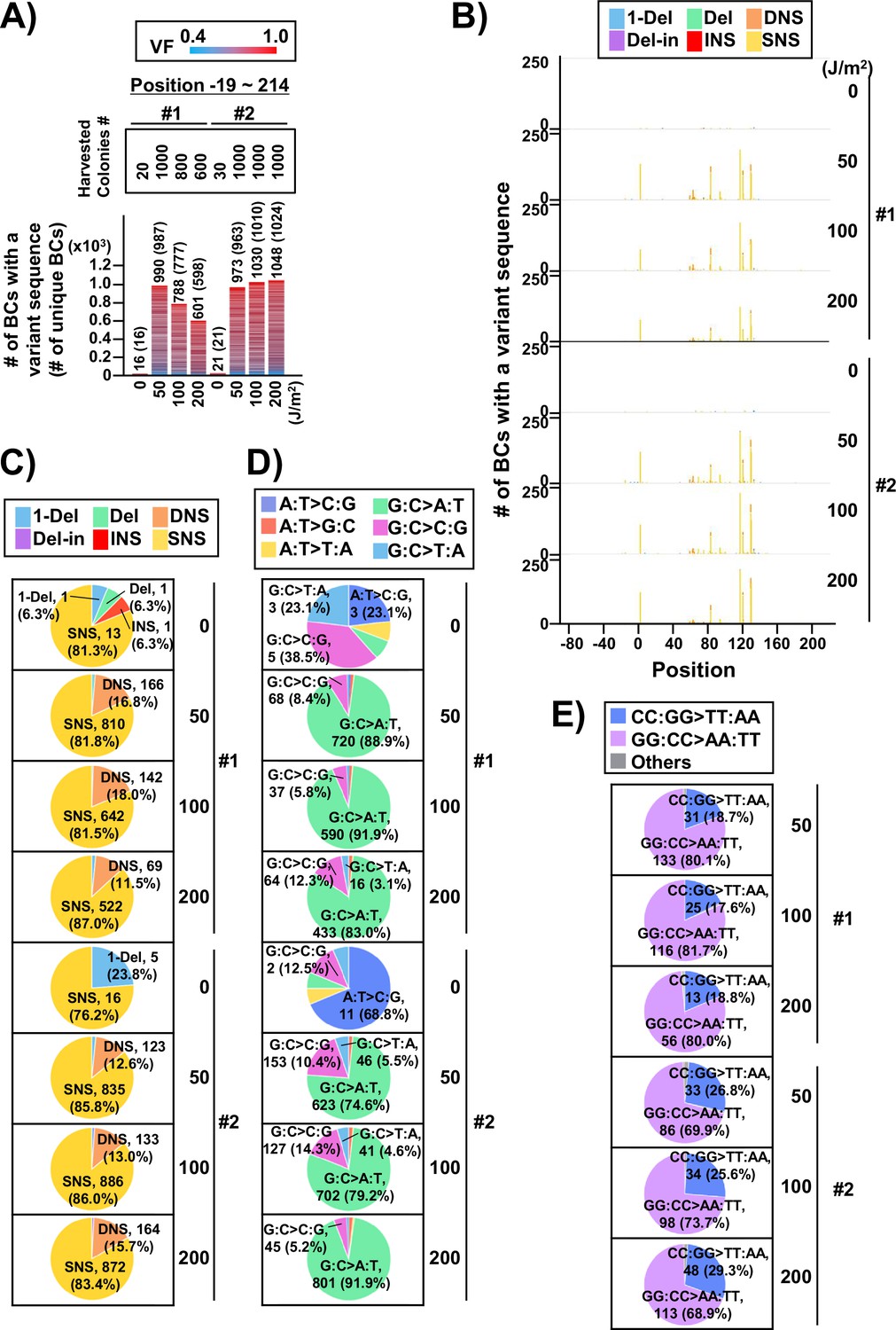
Development of a versatile high-throughput mutagenesis assay with multiplexed short-read NGS using DNA-barcoded supF shuttle vector library amplified in E. coli

aiGeneR 1.0: An Artificial Intelligence Technique for the Revelation of Informative and Antibiotic Resistant Genes in Escherichia coli

A simplified whole-genome sequencing library preparation workflow

PDF] Summarizing and correcting the GC content bias in high-throughput sequencing

Quality Control Solutions for Next-Gen Sequencing Workflows
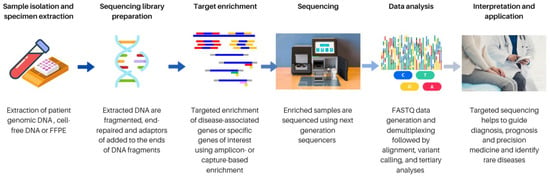
Cells, Free Full-Text
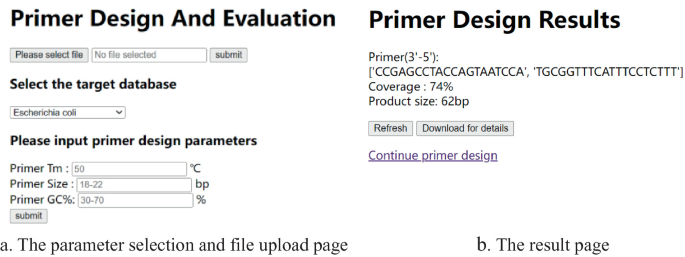
An Efficient and User-Friendly Software for PCR Primer Design for Detection of Highly Variable Bacteria

Identifying the best PCR enzyme for library amplification in NGS

Mapping and SNP Calling Tutorial